
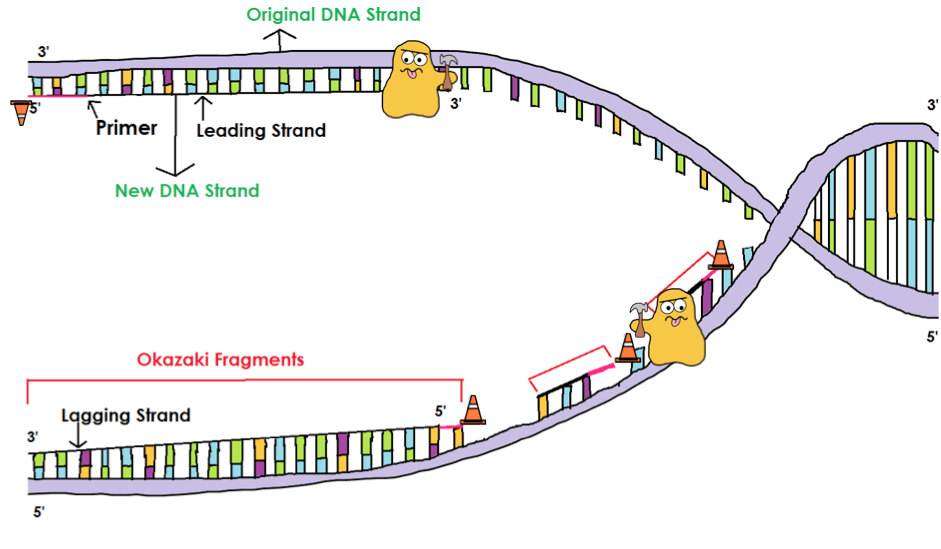
Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. DNA replication origin activation in space and time. DNA replication stress as a hallmark of cancer. The use of Ok-seq to interrogate genome-wide replication fork initiation and termination efficiencies can be applied to all unperturbed, asynchronously growing mammalian cells or under conditions of replication stress, and the assay can be performed in less than 2 weeks. Biotinylated Okazaki fragments are then captured on streptavidin beads and ligated to Illumina adapters before library preparation for Illumina sequencing. After size fractionation on a sucrose gradient, Okazaki fragments are concentrated and purified before click chemistry is used to tag the EdU label with a biotin conjugate that is cleavable under mild conditions. Briefly, cells are pulsed with 5-ethynyl-2′-deoxyuridine (EdU) to label newly synthesized DNA, and collected for DNA extraction.
Here we describe a detailed protocol for isolating and sequencing Okazaki fragments from asynchronously growing mammalian cells, termed Okazaki fragment sequencing (Ok-seq), for the purpose of quantitatively determining replication initiation and termination frequencies around specific genomic loci by meta-analyses. The ability to monitor DNA replication fork directionality at the genome-wide scale is paramount for a greater understanding of how genetic and environmental perturbations can impact replication dynamics in human cells.


 0 kommentar(er)
0 kommentar(er)
